

Long primers for RAPD mapping and fingerprinting of grape and pear
Guang-Ning Ye, Minou Hemmat, Muhammad A. Lodhi, Norman F. Weeden and Bruce I. Reisch
BioTechniques 20:368-371 (1996)
Department of Horticultural Sciences, New York State Agricultural Experiment Station, Cornell University, Geneva, New York 14456, USA
Key words: Polymerase chain reaction, Vitis, Pyrus
The RAPD (random amplified polymorphic DNA) technique (10, 12) has been widely used in plants for the construction of genetic maps in species such as Arabidopsis (9), bananas (4) and slash pine (7), and for genotype identification and taxonomic studies (2, 5). RAPD markers are detected by the use of short oligonucleotides of arbitrary sequence as primers for the amplification of segments of the target genome. Generally, 10-mer primers with 50-80% G+C content are preferred (8). However, complex banding patterns were also generated with primers as short as 5 bases (1). There are few reports on the use of long primers (over 12 bases) (10, 11). The potential value of long primers (17-24 bases) for generating RAPD polymorphisms was investigated in this study. We compared the use of both short and long primers in RAPD assays of two plant species: grape and pear.
DNA was extracted from leaves according to the methods of Lodhi et al. (6) for grape, and of Doyle and Doyle (3) for pear. RAPD amplification was performed in a reaction volume of 25 µl containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 2 mM MgCl2, 0.1% Triton X-100, 120 µM each of dNTPs, 0.4 µM primer, 100-200 ng genomic DNA and 0.5 unit of Taq DNA polymerase. Amplifications were performed on a PTC-100 thermocycler (MJ Research Inc., Watertown, MA, USA) for 1) 35 cycles of 30 sec at 94ûC, 1 min at 35ûC and 1 min 45 sec at 72ûC, followed by an 8 min extension at 72û for grape, or 2) 40 cycles of 1 min at 94ûC, 2 min at 35ûC and 2 min at 72ûC, followed by an 8 min extension at 72û for pear. Primers (10-mers) were purchased from Operon Technologies (Alameda, CA, USA), National BioSciences (Plymouth, MN, USA), Genosys Biotechnologies, Inc. (The Woodlands, TX, USA), University of British Columbia (Canada) and the New York State Center for Advanced Technology in Biotechnology (Cornell University, Ithaca, NY, USA). Long (17- to 24-mer) primers were provided by Dr. Sheng-Zhi Pang (Dept. of Plant Pathology, Cornell University, Geneva, NY, USA).
Typical gels displaying the amplification products generated from grape or pear DNA using long primers are shown in Fig. 1. In general, the long primers generated more DNA fragments, a wider range of DNA fragment sizes (typically 0.2-2 kb vs. 0.3-1.3 kb for 10-mer primers) and a greater number of polymorphic fragments per primer (Table 1). In grape genetic mapping studies, the number of polymorphic bands per 10-mer primer (4.8) is overestimated because the 77 primers used were the best among 310 surveyed, about 10% of which revealed few or no polymorphisms. When the data for all 310 primers was analyzed and compared with unselected long primers (Table 1), there was an even greater difference (2.9-fold) between the number of polymorphic bands generated with short primers vs. long primers. The results from studies in pear genetic mapping and grape variety fingerprinting showed the same trend (Table 1). Data from these two studies were generated with 10-mer primers which had been selected from a larger group.
No significant correlation was observed between primer length and number of polymorphic fragments per primer within the long primer group (Table 2), suggesting that increasing primer length above 20 bases may not be an efficient method for exposing more polymorphism. Generally, 10-mer primers with 50-80% G+C content are preferred (8) in RAPD analyses. As the length of a primer increases, the genomic target sites should decrease because there is less chance of finding perfect or near perfect homologies between the target sites and a longer primer. We observed, however, that long primers yielded more polymorphic bands than the short 10-mer primers, which is consistent with the observation on poplar (2). Long primers have also been used in genomic fingerprinting (10, 11). However, no data were presented comparing RAPD results between 10-mer primers and longer primers (2, 10, 11). It is not clear exactly why the long primers produced more polymorphic bands. It should be pointed out, however, that the long primers used in this study have lower G+C content (from 29% to 55%) compared to commonly used 10-mer primers, those used by Welsh and McClelland (10, 11) (45-61%) and those by Castiglione et al. (2) (45-60%), yet they were able to generate more fragments than 10-mer primers. It is not clear in our study whether the increased number of bands produced by long primers was due to lower G+C content, primer length, or a combination of both. Future experiments should be designed to determine the effects of primer length and G+C content upon RAPD polymorphisms in a variety of genera. If the generation of more fragments is due to primer length, one possibility could be that the extra bases at the 5' end anneal to the templates in a way that either helps the 3' bases anneal to the template to start a new template-primer complex or stabilizes the unstable existing template-primer complex. Because most of the primers have less than 50% G+C content, intergenic or repetitive DNA regions may be preferentially targeted, which could be useful in mapping telomere and centromere regions that are otherwise not as easily accessible with 10-mer primers. Sequencing of the amplified fragments or probing of the genomic DNA with the amplified fragments would help to determine the nature of amplified products and the mechanism(s) underlying the amplification.
The higher cost for synthesizing long primers can be justified by the greater number of polymorphic bands obtained. Long primers might also be available gratis from colleagues in a large research facility. To generate the same number of polymorphic bands, reactions with 10-mer primers cost up to three times as much in materials and labor as reactions with long primers.
Acknowledgments
We are grateful to Dr. Sheng-Zhi Pang for providing the long primers and Dr. Richard Bell for supplying pear germplasm. We thank Sheri Woo and Tim Johnson for technical assistance, and Drs. Thomas Björkman and John Norelli for critical review of this manuscript. This research was supported by Grant No. US1888-90 from BARD, the US-Israel Binational Agricultural Research and Development Fund, by the New York Wine and Grape Foundation, and by CSRS specific agreement No. 58-1931-2-046.
References
1.Caetano-Anolles, G., B.J. Bassam and P.M. Gresshoff. 1991. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Bio/Technology. 9:553-557.
2.Castiglione, S., G. Wang, G. Damiani, C. Bandi, S. Bisoffi and F. Sala. 1993. RAPD fingerprints for identification and for taxonomic studies of elite poplar (Populus spp.) clones. Theor. Appl. Genet. 87:54-59.
3.Doyle, J.J. and J.L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12:13-15.
4.Faure, S., J.L. Noyer, J.P. Horry, F. Bakry, C. Lanaud and D.G. de Leon 1993. A molecular marker-based linkage map of diploid bananas (Musa acuminata). Theor. Appl. Genet. 87:517-526.
5.Hu, J. and C.F. Quiros. 1991. Identification of broccoli and cauliflower cultivars with RAPD markers. Plant Cell Rep. 10:505-511.
6.Lodhi, M.A., G.-N. Ye, N.F. Weeden and B.I. Reisch. 1994. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Pl. Mol. Biol. Rep. 12:25-32.
7.Nelson, C.D., W.L. Nance and R.L. Doudrick. 1993. A partial genetic linkage map of slash pine (Pinus elliottii Engelm. var. elliottii) based on random amplified polymorphic DNAs. Theor. Appl. Genet. 87:145-151.
8.Rafalski, J.A., S.V. Tingey and J.G.K. William. 1991. RAPD markers-a new technology for genetic mapping and plant breeding. AgBiotech News and Information. 3:645-648.
9.Reiter, R.S., J.G.K. Williams, K.A. Feldmann, J.A. Rafalski, S.V. Tingey and P.A. Scolnik. 1992. Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc. Natl. Acad. Sci. U.S.A. 89:1477-1481.
10.Welsh, J. and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218.
11.Welsh, J. and M. McClelland. 1991. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucl. Acids Res. 19:5275-5279.
12.Williams, J.G.K., A.R. Kubelik, K.J. Livak, J.A. Rafalski and S.V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535.
|
Table 1. Comparison of 10-mer and 17- to 24-mer primers
in genetic mapping and fingerprinting of grape and pear |
||
| 10-mer Primers | 17- to 24-mer Primers | |
| Pear genetic mapping population | ||
No. of primers |
52 | 7 |
Mean no. DNA fragments/primer |
10.5 | 14.3* |
Mean no. polymorphic fragments/primer |
6.2 | 8.4* |
| Grape genetic mapping population | ||
No. of primers |
77 (310)a | 13 (20) |
Mean no. DNA fragments/primer |
12.1 (5.9) | 15.8* (14.9) |
Mean no. polymorphic fragments/primer |
4.8 (2.6) | 6.7* (7.6) |
| Grape variety fingerprinting | ||
No. of primers |
43 | 11 |
Mean no. DNA fragments/primer |
5.6 | 11.8* |
|
3.0 | 4.5* |
| a The numbers in parentheses represent the combined data for primers tested on parents only, as well as those used with the progeny population. Numbers not in parentheses represent data from primers chosen for analysis with a progeny population. | ||
| * Significantly different at p<0.001 according to a one-tailed t-test. | ||
| Table 2. The effect of primer length (17
to 24 bases) on number of RAPD fragments and number of polymorphic fragments
per primer |
|||
|
Primer Length (Bases)
|
No. of Primers Tested a
|
Average total no. of Fragments/Primer
|
Average no. of polymorphic Fragments/Primer
|
|
17 |
3
|
12
|
6.7
|
| 18 |
9
|
18
|
8.8
|
| 19 |
3
|
12
|
4.3
|
| 21 |
3
|
13
|
6.7
|
| 22 |
1
|
17
|
7.0
|
| 24 |
1
|
18
|
7.0
|
| a The data include results from primers used in grape and pear. Thirteen primers were used in grape and seven in pear (see Table 1), of which 5 were the same primers. Where the same primers were used in grape and pear they were counted twice in the computation. | |||
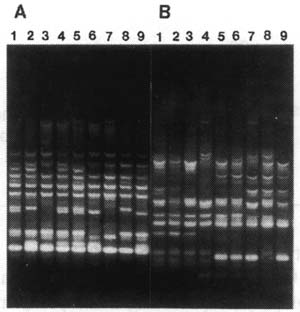
Figure 1. RAPD profiles of grape DNA generated with the 21-mer GY169 (CTAAGCTGCTTTTGTTTGAGC) (panel A) and pear DNA with the 18-mer GY107 (GTTCAGGGCTGTTTATAG) (panel B). Each panel includes DNA from 9 individuals from the crosses Horizon x Ill. 547-1 (Panel A) and Bartlett x NY10353 (Panel B), respectively. The amplification products were separated by electrophoresis in 2% agarose gels (1% agarose/1% NuSieve GTG agarose, FMC Corp., Rockland, Maine, USA), visualized by staining with ethidium bromide, and photographed on a transilluminator using Polaroid Type 55 film.
Back to Grape Genetics Links.